BIOLOGICAL BUFFERS
BIOLOGICAL BUFFERS
Biological Buffers
The pKa of a buffer is commonly perceived as the pH of the said buffer when the concentrations of the two buffering species are equal, and where the maximum buffering capacity is achieved. However, it is often forgotten, that when defined as above, pKa depends on buffer concentration and temperature. To avoid this problem the concept of “thermodynamic” pKa0 was introduced. pKa0 is pKa of the buffer at infinite dilution (buffer concentration=0) and 25oC. Thus, pKa0 is a true constant specific for a given buffer.
Note that, depending on the nature of the buffer, the pH (and pKa) of the buffer solution may increase or decrease upon dilution, and this effect may be significant. Additionally, small changes in temperature can also cause noticeable changes in the pH of the buffer solution. For the buffers shown on the brown background, the buffers' concentrations have especially strong influfluences on the buffers' pKa values.
For your convenience, the biological buffers table contain values of pKa0, d(pKa0)/dt at 298.25 K, and a calculator that allows you to estimate pKa values of each buffer at temperatures form 3oC to 37oC, and concentrations from 1 to 500mM for "white background" buffers and 1 to 130mM for "brown background" buffers.
To use the calculator, enter the buffer's concentration and temperature, then click on the corresponding button.**
| pKa0 (25 oC) | d(pKa)/dt ΔH0, kJ/mol |
Calculate pKa at given concentration and temperature |
Buffer name/dissociation type | Dissociation step | UV limit |
Notes |
| 1.92 | -0.0006 1.1 |
mM oC pKa |
Maleic acid H2L = H+ + HL-1 |
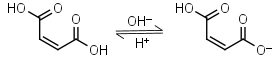 |
270nm | Other pKa: 6.23. Can react with strong nucleophiles (e.g. thiols) by Michael addition. |
| 2.148 | +0.0047 -8.0 |
mM oC pKa |
Phosphoric acid H3L = H+ + H2L-1 |
H3PO4 = H+ + H2PO4-1 | Clear | Other pKa: 7.198, 12.35. Stabilizes many enzymes. Precipitates bivalent cations. |
| 3.128 | -0.0024 4.07 |
mM oC pKa |
Citric acid H3L = H+ + H2L-1 |
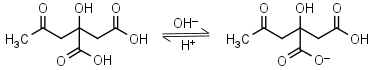 |
Clear | Other pKa: 4.761, 6.396. Binds to some proteins. Forms complexes with many metals. |
| 3.40 | mM 25oC pKa |
Malic acid H2L = H+ + HL-1 |
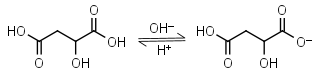 |
Clear | Other pKa: 5.11. Is chiral. Forms complexes with some metals. | |
| 3.75 | mM 25oC pKa |
Formic acid HL = H+ + L-1 |
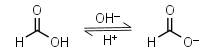 |
Clear | Slightly volatile, usually is sold containing 5-10% of water - difficult to dose in exact amounts. | |
| 3.86 | mM 25oC pKa |
Lactic acid HL = H+ + L-1 |
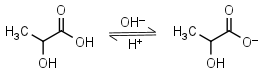 |
Clear | Is chiral. Usually is sold as ~85% solution in water - difficult to dose in exact amounts. | |
| 4.207 | -0.0018 3.0 |
mM oC pKa |
Succinic acid H2L = H+ + HL-1 |
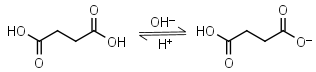 |
Clear | Other pKa: 5.636. |
| 4.756 | +0.0002 -0.41 |
mM oC pKa |
Acetic acid HL = H+ + L-1 |
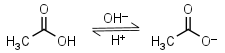 |
Clear | Volatile. |
| 5.03 (at 20oC) | mM 20oC pKa |
Pivalic (trimethylacetic) acid HL = H+ + L-1 |
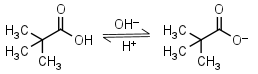 |
Clear | Volatile, has bad odor and relatively low solubility in water. | |
| 5.11 | mM 25oC pKa |
Malic acid HL-1 = H+ + L-2 |
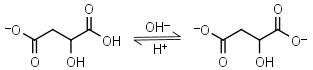 |
Clear | Other pKa: 3.40. Is chiral. Forms complexes with some metals. | |
| 5.23 | -0.014 | mM oC pKa |
Pyridine HL+ = H+ + L |
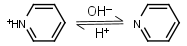 |
275nm | Volatile and toxic. |
| 5.333 | -0.018 31.11 |
mM oC pKa |
Piperazine H2L+2 = H+ + HL+ |
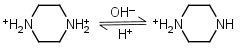 |
Clear | Other pKa: 9.731. |
| 5.39 (at 20oC) | mM 20oC pKa |
Picolinic acid HL± = H+ + L-1 |
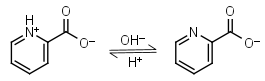 |
Other pKa: 0.99 at 20oC. | ||
| 5.636 | +0.0002 -0.5 |
mM oC pKa |
Succinic acid HL-1 = H+ + L-2 |
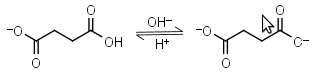 |
Clear | Other pKa: 4.207. |
| 6.05 | -0.017 29.5 |
mM oC pKa |
L-Histidine H2L+ = H+ + HL± |
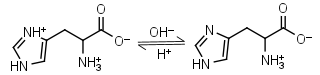 |
235nm | Other pKa: 1.80, 9.34. Chiral. Forms complexes with Me2+, forms complexes with itself. It is a primary amine, and therefore can form Schiff’s bases with aldehydes/ ketones. |
| 6.20 | -0.0087 14.8 |
mM oC pKa |
MES HL± = H+ + L-1 |
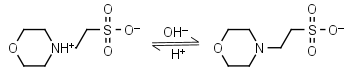 |
Clear | Other pKa: <3. Weakly binds Ca, Mg, Mn. Negligible binding with Cu(II).1,2 |
| 6.484 | -0.017 28.4 |
mM oC pKa |
Bis-tris HL+ = H+ + L |
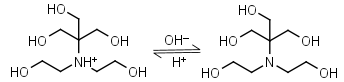 |
Clear | Binds Ca(II), Sr(II), Co(II), Ni(II), Cu(II), Zn(II), Cd(II), Pb(II); weakly binds Mg(II), Ba(II), Mn(II).11 |
| 6.65 | -0.03 |
mM oC pKa |
Bis-tris propane H2L+2 = H+ + HL+ |
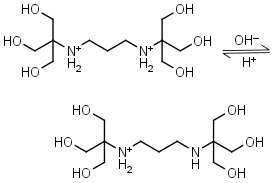 |
Clear | Other pKa: 9.11. |
| 6.844 | -0.0072 12.23 |
mM oC pKa |
ADA HL-1 = H+ + L-2 |
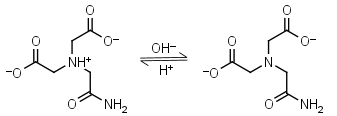 |
260nm | Other pKa: 1.59, 2.48. Binds Cu(II), Co(II), Zn(II), Mn(II), Ni(II), Ca(II), weakly binds Mg(II).3,4 Free acid is poorly soluble in water. |
| 6.847 | -0.018 30.43 |
mM oC pKa |
ACES HL± = H+ + L-1 |
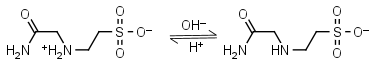 |
230nm | Other pKa: <3. Binds Cu(II), Co(II), Zn(II), Nil(II), weak binding to Ca(II), Mg(II), negligible binding to Mn(II).1,5 |
| 6.90 | -0.015 25.0 |
mM oC pKa |
MOPSO HL± = H+ + L-1 |
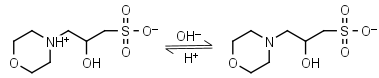 |
Clear | Other pKa: <2. Chiral, some metal binding. |
| 6.960 | -0.0072 12.3 |
mM oC pKa |
PIPES (HL±)-1 = H+ + L-2 |
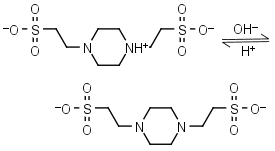 |
Clear | Other pKa: <3. Negligible binding to metals.1,2 Can form radicals, should be avoided in studies of oxidative compounds and redox processes in biochemistry.6,7 Free acid is poorly soluble in water. |
| 6.993 | -0.021 36.64 |
mM oC pKa |
Imidazole HL+ = H+ + L |
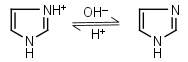 |
235nm | Forms complexes with Me2+, and forms complexes with histidine. Strongly nucleophilic, catalyzes wide range of chemical transformations. |
| 7.184 | -0.012 21.1 |
mM oC pKa |
MOPS HL± = H+ + L-1 |
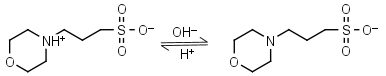 |
Clear | Other pKa: <2. Negligible binding to metals.2 Partially degrades on autoclaving in the presence of glucose. |
| 7.187 | -0.014 24.25 |
mM oC pKa |
BES HL± = H+ + L-1 |
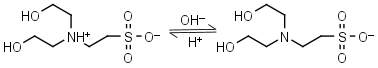 |
Clear | Other pKa: <3. Binds Cu(II), negligible binding to Ca(II), Mg(II) and Mn(II).1 |
| 7.198 | -0.0022 3.6 |
mM oC pKa |
Phosphoric acid H2L-1 = H+ + HL-2 |
H2PO4-1 = H+ + HPO4-2 | Clear | Other pKa: 2.148, 12.35. Stabilizes many enzymes. Precipitates bivalent cations. |
| 7.50 | -0.019 32.13 |
mM oC pKa |
TES HL± = H+ + L-1 |
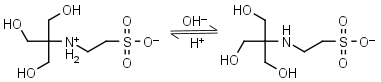 |
Clear | Other pKa: <3. Binds Cu(II), Co(II), Zn(II), Mn(II). Negligible binding to Ca(II), Mg(II) and Mn(II).1,8 |
| 7.564 | -0.012 20.4 |
mM oC pKa |
HEPES HL± = H+ + L-1 |
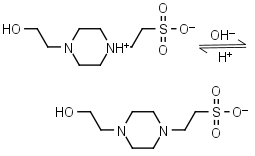 |
Clear | Other pKa: <3. Negligible binding to Ca(II), Mg(II) and Mn(II).1 Is oxidized by Cu(II).9 Can form radicals, should be avoided in studies of redox processes in biochemistry.6,7 |
| 7.576 | -0.018 30.18 |
mM oC pKa |
DIPSO HL± = H+ + L-1 |
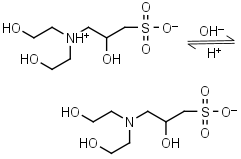 |
Clear | Other pKa: <2. Chiral. Binds Co(II), Ni(II).10 |
| 7.635 | -0.023 39.09 |
mM oC pKa |
TAPSO HL± = H+ + L-1 |
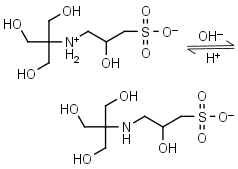 |
Clear | Other pKa: <2. Chiral. Binds Co(II), Ni(II).10 |
| 7.762 | -0.020 33.6 |
mM oC pKa |
TEA (Triethanolamine) HL+ = H+ + L |
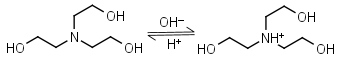 |
Clear | Binds Co(II), Ni(II), Cu(II), Zn(II), Cd(II).11 Can form radicals in the presence of strong oxidants, exercise caution during studies of redox processes. |
| 7.77 | -0.016 27.4 |
mM oC pKa |
N-Ethylmorpholine HL+ = H+ + L |
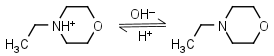 |
Clear | |
| 7.85 | -0.013 |
mM oC pKa |
POPSO (HL±)-1 = H+ + L-2 |
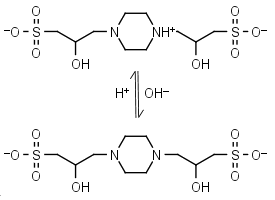 |
Clear | Other pKa: <2. Chiral, is a mixture of two diastereomers. Can form radicals, should be avoided in studies of redox processes in biochemistry.6,7 Free acid is poorly soluble in water. |
| 7.957 | -0.013 21.3 |
mM oC pKa |
EPPS, HEPPS HL± = H+ + L-1 |
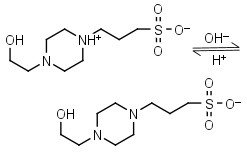 |
Clear | Other pKa: <2. Binds metals. Can form radicals, should be avoided in studies of redox processes in biochemistry.6,7 |
| 7.94 | -0.014 23.70 |
mM oC pKa |
HEPPSO HL± = H+ + L-1 |
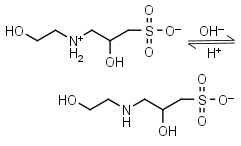 |
Clear | Other pKa: <2. Chiral. |
| 8.072 | -0.028 47.45 |
mM oC pKa |
Tris HL+ = H+ + L |
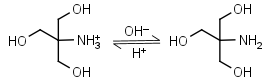 |
Clear | Binds Cu(II), Ni(II).11,12 Binds Co(II), Zn(II), Cd(II), Pb(II); weakly binds Ca(II), Mg(II), Ba(II), Mn(II).11 It is a primary amine, and therefore can form Schiff’s bases with aldehydes/ ketones. Inactivates DEPC. Is involved in some enzymatic reactions (e.g. alkaline phosphatase). |
| 8.135 | -0.018 31.37 |
mM oC pKa |
Tricine HL± = H+ + L-1 |
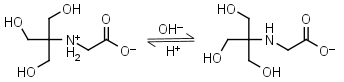 |
Clear | Other pKa: 2.023. Binds Cu(II), Co(II), Zn(II), Ni(II), Cd(II), Pb(II) Ca(II), Mg(II) and Mn(II).1,4 Is photooxidized by flavines. |
| 8.265 | -0.026 43.40 |
mM oC pKa |
Glycylglycine HL± = H+ + L-1 |
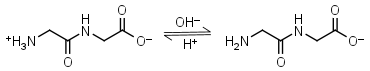 |
Clear | Other pKa: 3.14. Binds Cu(II) Mn(II), weakly binds Ca(II) and Mg(II).1 Is a primary amine, therefore it can form Schiff’s bases with aldehydes/ ketones. |
| 8.334 | -0.016 26.34 |
mM oC pKa |
Bicine HL± = H+ + L-1 |
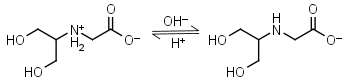 |
Clear | Other pKa: 2.0. Binds Cu(II), Co(II), Zn(II), Mn(II), Ca(II), Mg(II).1,8 Is slowly oxidized by ferricyanide. |
| 8.44 | -0.024 40.4 |
mM oC pKa |
TAPS HL± = H+ + L-1 |
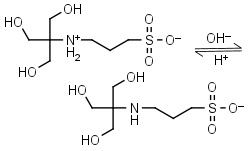 |
Clear | Other pKa: <2. Binds Co(II), Ni(II).10 |
| 8.50 | -0.022 |
mM oC pKa |
Morpholine HL+ = H+ + L |
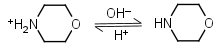 |
Clear | |
| 8.54 | -0.028 |
mM oC pKa |
N-Methyldiethanolamine HL+ = H+ + L |
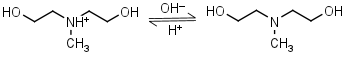 |
Clear | |
| 8.801 | -0.029 49.85 |
mM oC pKa |
AMPD (2-amino-2-methyl-1,3-propanediol) HL+ = H+ + L |
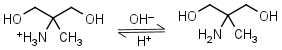 |
Clear | It is a primary amine, and therefore can form Schiff’s bases with aldehydes/ ketones. |
| 8.883 | -0.026 42.08 |
mM oC pKa |
Diethanolamine HL+ = H+ + L |
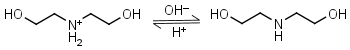 |
Clear | Binds Ni(II), Cu(II), Zn(II), Cd(II).11 |
| 9.138 | -0.025 43.19 |
mM oC pKa |
AMPSO HL± = H+ + L-1 |
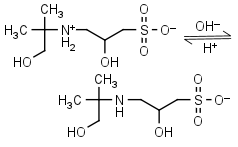 |
Clear | Other pKa: <2. Binds Co(II), Ni(II).10 Is chiral. |
| 9.237 | -0.008 13.8 |
mM oC pKa |
Boric acid HL = H+ + L-1 |
H3BO3 = H+ + H2BO3-1 | Clear | Forms covalent complexes with mono- and oligosaccharides, ribose subunits of nucleic acids, pyridine nucleotides. |
| 9.43 | -0.023 39.55 |
mM oC pKa |
CHES HL± = H+ + L-1 |
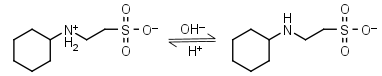 |
Clear | Other pKa: <3. |
| 9.780 | -0.026 44.2 |
mM oC pKa |
Glycine HL± = H+ + L-1 |
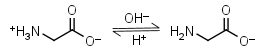 |
Clear | Other pKa: 2.351. Interferes with Bradford protein assay. Is a primary amine, therefore it can form Schiff’s bases with aldehydes/ ketones. |
| 9.825 | -0.027 46.67 |
mM oC pKa |
CAPSO HL± = H+ + L-1 |
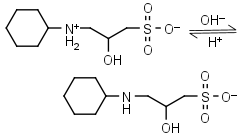 |
Clear | Other pKa: <2. Is chiral. |
| 9.498 | -0.030 50.52 |
mM oC pKa |
Ethanolamine HL+ = H+ + L |
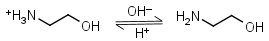 |
Clear | Binds Co(II), Ni(II), Cu(II), Zn(II), Cd(II); weakly binds Mn(II).11 It is a primary amine, and therefore can form Schiff’s bases with aldehydes/ ketones. |
| 9.694 | -0.032 54.05 |
mM oC pKa |
AMP (2-amino-2-methyl-1-propanol) HL+ = H+ + L |
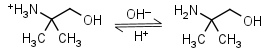 |
Clear | It is a primary amine, and therefore can form Schiff’s bases with aldehydes/ ketones. |
| 9.731 | -0.025 42.89 |
mM oC pKa |
Piperazine HL+ = H+ + L |
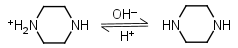 |
Clear | Other pKa: 5.333. |
| 10.499 | -0.028 48.1 |
mM oC pKa |
CAPS HL± = H+ + L-1 |
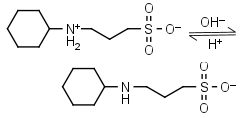 |
Clear | Other pKa: <2. |
| 10.55 | -0.026 |
mM oC pKa |
1,3-Diaminopropane HL+ = H+ + L |
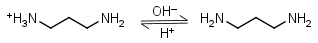 |
Clear | Other pKa: 8.88. Forms strong complexes with many metals. It is a primary amine, and therefore can form Schiff’s bases with aldehydes/ ketones. |
| 10.7 | mM 25oC pKa |
CABS HL± = H+ + L-1 |
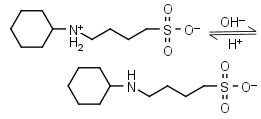 |
Clear | Other pKa: <2. | |
| 11.123 | -0.031 |
mM oC pKa |
Piperidine HL+ = H+ + L |
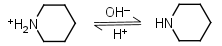 |
Clear |
*
Significant deviations exist in the reported values of pKa and other thermodynamic constants of most common buffers due to them being determined by methods of different accuracy. Additionally, many online resources provide pKa values of biological buffers at unspecified or wrongly specified ionic strengths. We attempted to provide the most consistent data available. pKa0, d(pKa)/dt and ΔH0 are compiled mostly from
- CRC Handbook of Chemistry & Physics, 93th edition: Dissociation Constants of Organic Acids and Bases.
- Goldberg, R. N., Kishore, N., Lennen, R. M. J. Phys. Chem. Ref. Data, 31, 2002, 231-370, as well as some other original publications.
If you have have experimentally observed significantly different values, please report them to support@reachdevices.com.
**Temperature dependence of pKa is presumed to be linear.
White background buffers:
- for concentrations 1 to 200mM, the Debye-Hückel model is used, and the resulting pKa is presented in brown font.
- for concentrations 201 to 500mM, the Davies model is used, and the resulting pKa is presented in blue font.
Light brown background buffers:
- for concentrations 1 to 50mM, the Debye-Hückel model is used, and the resulting pKa is presented in brown font.
- for concentrations 51 to 130mM, the Davies model is used, and the resulting pKa is presented in blue font.
For a detailed explanation of pKa vs pKa0, and formulas used in the calculator, click on this link.
1 - Good, N. E., Winget, G. D., Winter, W., Connolly, T. N., Izawa, S., Singh. R. M. M. Biochemistry, 1966, 5, 467-477.
2 - Kandegedara, A., Rorabacher, D. B. Anal. Chem., 1999, 71, 3140-3144.
3 - Lance, E. A., Rhodes, C.W. , Nakon, R. Anal. Biochem., 1983, 133, 492-501.
4 - Nakon, R. Anal. Biochem., 1979, 95, 527-532.
5 - Pope, J. M., Stevens, P. R., Angotti, M. T., Nakon. R. Anal. Biochem., 1980, 103, 214-221.
6 - Grady, J. K., Chasteen, N. D., Harris. D. C., Anal. Biochem., 1988, 173, 111-115.
7 - Kirsch, M., Lomonosova, E. E., Korth, H.-G., Sustmann, R. de Groot, H. The Journal of Biological Chemistry, 1998, 273, 12716-12724.
8 - Nakon, R., Krishnamoorthy. C. R. Science, 1983, 221, 749-750.
9 - Hegetschweiler, K., Saltman, P. Inorg. Chem., 1986, 25, 107-109.
10 - Machado, C. M. M., Gameiro, P., Soares, H. M. V. M. J. Solution Chem., 2008, 37, 603-617.
11 - Scheller, K. H., Abel, T. H., Polanyi, P. E., Wenk, P. K., Fischer, B. E., Sigel. H. Eur. J. Biochem., 1980, 107, 455-466.
12 - Bai, K. -S., Martell. A. E., J. Inorg. Nucl. Chem., 1969, 31, 1697–1707.
REACH Devices, LLC.
6525 Gunpark Drive, Suite 370-179, Boulder, CO 80301
Call: (720) 288-5722
Inquiries: support@reachdevices.com
Ⓒ 2010-2025 REACH Devices, LLC. All rights reserved.